Resource | Mollie's Blog No 17
Meet the Scientists
Since my previous blog, I have been working to answer the unexpected question that arose from the last set of dephosphorylation assays I had performed using whole cell extracts: where is PP1 located and active in the cell? We had hypothesized that, due to the location of its substrate (phosphorylated ubiquitin, pUb, at the mitochondria), it was likely PP1 may be tethered to the mitochondrial membrane. This would enable PP1 to be in close proximity with pUb and easily carry out its dephosphorylation activity. This is, however, just supposition and needed to be clarified experimentally.
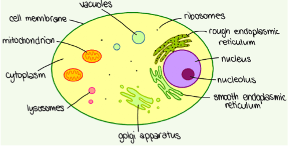
To investigate this, I proceeded to separate the proteins of the whole cell extract into 3 distinct groups: a nuclear fraction, containing all the proteins found in or at the nucleus; a mitochondrial fraction, containing all the proteins found in or at the mitochondria and finally a cytosolic fraction, this contains all the proteins that are not associated with a particular membrane and subsequently are free to move throughout the cell. It is important to note that some proteins spend some time free floating but in response to certain stimuli may move to a particular membrane such as the rough or smooth endoplasmic reticulum, mitochondria, lysosomes or golgi apparatus in the diagram. Here, the protein may carry out a certain function – this behaviour could be what is occurring with PP1 in response to the accumulation of pUb at the mitochondria.
Intriguingly, after a couple of repetitions of the experiment, the dephosphorylation activity against pUb is appearing in the cytosolic fraction. This indicates that PP1 is not permanently tethered to the mitochondrial membrane as we proposed, instead these results suggest that PP1 is a free protein that can move throughout the cytosol. These findings are really interesting and suggest some mechanism is in place (such as the recruitment of a very specific subunit) that can guide PP1 to the mitochondria to allow it to act on pUb.
These findings also open up the possibility to make use of another very illuminating technique – gel filtration. I will explain the mechanism of gel filtration in a later blog but briefly, this technique could allow us to identify the binding partners of PP1 that facilitate its activity against pUb. This data would hopefully back up the genetic data from the siRNA screen, adding greater significance to any conclusions we may draw. It is likely that this work will not be completed before I finish my placement, however, I am both extremely grateful and excited to be laying the foundations for this particular line of enquiry into the PINK1/Parkin pathway.
Aside from lab work, this month MRC PPU are undertaking a 30-day challenge to raise money for Parkinson’s Disease research. I am organising a unit-wide challenge to cover as much distance as possible over the month of June by encouraging staff in the lab to get out and run, cycle, swim or hike as much as possible this month. Staff are also encouraged to switch their commute to a greener alternative (cycling or walking rather than driving). I am compiling our total miles and the total elevation gained and reporting our progress each week. I initially set a target to complete the same distance as the North Coast 500, yet, just two weeks into our challenge and we have already exceeded this distance – together completing 565 miles with the equivalent elevation gain of climbing Ben Nevis more than 9 times. If you would like to share our JustGiving page with friends and family to help us raise money to support the fantastic work being carried out into Parkinson’s Disease here is our team link: https://www.justgiving.com/team/NorthCoast500?utm_source=facebook&utm_medium=team&utm_content=NorthCoast500&utm_campaign=pfp-share&utm_term=40cbbc9ef2764453ac5c4024de2952e3&fbclid=IwAR0aUN9mKayLPadp-U7Oua6LL-S1JEO2cE5MCgVFDLO2XNYaPDHBuyCmGPc
Many thanks in advance and hopefully in my next blog I can blow you away with the distance we manage to cover by the end of the month.
